Neural Networks
Definitions
NNs are computing systems inspired by the biological neural networks that constitute human brains.
They are based on a collection of connected notes called artificial neurons.
Each connection, akin to synapses in a biological brain, can transmit a signal to other neurons, therefore processing any information given as inputs to produce a final signal as output.
Training neural networks can be done with algorithms such as Backpropagation.
Simple Neural Net
__init__method listing trainable parameters, i.e a weight vectorwith 2 elements and a single scalar bias value forwardmethod to formulate predictions for any set of given inputs- Adding a loss function: Reuse the MSE
- The NN could operate with any number of inputs
and outputs by representing: as a matrix as a vector
class SimpleNeuralNet():
def __init__(self, W, b):
self.W = W
self.b = b
# Add loss
self.loss = float("Inf")
def forward(self, x):
Z = np.matmul(x, self.W)
pred = Z + self.b
return pred
def MSE_loss(self, inputs, outputs):
outputs_re = outputs.reshape(-1, 1)
pred = self.forward(inputs)
losses = (pred - outputs_re) ** 2
self.loss = np.sum(losses) / output.shape[0]
return self.loss
simple_nn = SimpleNeuralNet(W = np.array([], []), b = np.ones(shape = (1,1)))
Shallow Neural Net
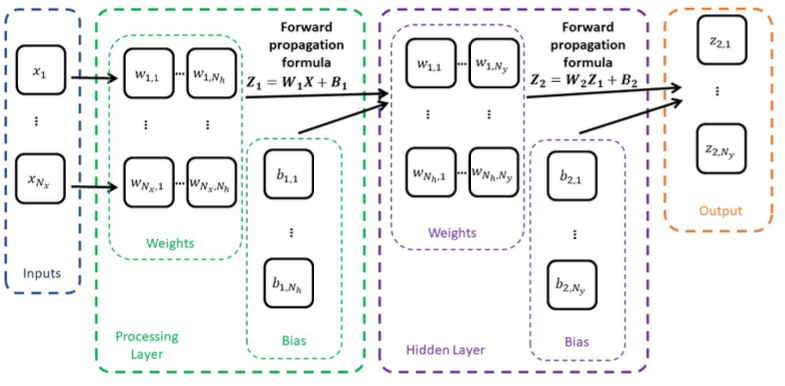
It will include two processing layers of
- First layer will receive inputs with dimensionality
and produce outputs with dimensionality . - Second layer called the hidden layer will receive inputs from previous layer (of the dimensionality
in this case) and produce outputs with dimensionality of outputs in our dataset - Hence, matrix
is 2D and is 1D
class ShallowNeuralNet():
def __init__(self, n_x, n_h, n_y):
# Network dimensions
self.n_x = n_x
self.n_h = n_h
self.n_y = n_y
# Weights and biases matrices
self.W1 = np.random.randn(n_x, n_h)*0.1
self.b1 = np.random.randn(1, n_h)*0.1
self.W2 = np.random.randn(n_h, n_y)*0.1
self.b2 = np.random.randn(1, n_y)*0.1
# Loss, initialized as infinity before first calculation is made
self.loss = float("Inf")
def forward(self, inputs):
# Wx + b operation for the first layer
Z1 = np.matmul(inputs, self.W1)
Z1_b = Z1 + self.b1
# Wx + b operation for the second layer
Z2 = np.matmul(Z1_b, self.W2)
Z2_b = Z2 + self.b2
return Z2_b
def MSE_loss(self, inputs, outputs):
# MSE loss function as before
outputs_re = outputs.reshape(-1, 1)
pred = self.forward(inputs)
losses = (pred - outputs_re)**2
self.loss = np.sum(losses)/outputs.shape[0]
return self.loss
We now need a training procedure since the weights and biases are currently randomly generated (See Backpropagation).
Training Procedure
- Backward Method - performs calculation of gradients in matrix form and parameter adjustments using GD update rules
- Forward Method - repeats backward method, reusing GD for-loop function until a maximal number of iterations is reached, or convergence is seen
i.e. the procedure:
- Forward pass on dataset samples
- Compute errors and loss function
- Use new loss value to compute gradients
- Adjust parameters with Backpropagation
- Repeat until convergence or max number of iterations
def train(self, inputs, outputs, N_max = 1000, alpha = 1e-5, delta = 1e-5, display = True):
# List of losses, starts with the current loss
self.losses_list = [self.loss]
# Repeat iterations
for iteration_number in range(1, N_max + 1):
# Backpropagate
self.backward(inputs, outputs, alpha)
new_loss = self.loss
# Update losses list
self.losses_list.append(new_loss)
# Display
if(display):
print("Iteration {} - Loss = {}".format(iteration_number, new_loss))
# Check for delta value and early stop criterion
difference = abs(self.losses_list[-1] - self.losses_list[-2])
if(difference < delta):
if(display):
print("Stopping early - loss evolution was less than delta.")
break
else:
# Else on for loop will execute if break did not trigger
if(display):
print("Stopping - Maximal number of iterations reached.")
def show_losses_over_training(self):
# Initialize matplotlib
fig, axs = plt.subplots(1, 2, figsize = (15, 5))
axs[0].plot(list(range(len(self.losses_list))), self.losses_list)
axs[0].set_xlabel("Iteration number")
axs[0].set_ylabel("Loss")
axs[1].plot(list(range(len(self.losses_list))), self.losses_list)
axs[1].set_xlabel("Iteration number")
axs[1].set_ylabel("Loss (in logarithmic scale)")
axs[1].set_yscale("log")
# Display
plt.show()
Symmetry
Initialising all parameters as identical constants (e.g. all zeros) is bad because of the tendency of all neurons to have the same weights and processes.
The lack of diversity leads to a lack of generalisation which prevents the NN from learning complex patterns.
This happens because all weights and baises had the same starting point. The backward process then updated the parameters identically and they end up keeping the same values over the course of training. Hence, we need random initialisation.
Symmetrical neural networks are vulnerable to Adversarial attacks
Model design
Constant initialisation is important to consider as it may lead to issues such as Vanishing gradients. Instead of zero/same constant initialisation, we need to use random starting values, or variants such as Xavier initialisation.
Selecting a proper learning rate is also important to tackle Exploding gradients.
Also, if our model only consists of two linear layers (as seen above), the boundary for our models is also linear in
Introducing non-linearity
Adding sigmoid operations after each linear operation in the forward method:
def forward(self, inputs):
# Wx + b operation for the first layer
Z1 = np.matmul(inputs, self.W1)
Z1_b = Z1 + self.b1
A1 = self.sigmoid(Z1_b)
# Wx + b operation for the second layer
Z2 = np.matmul(A1, self.W2)
Z2_b = Z2 + self.b2
y_pred = self.sigmoid(Z2_b)
return y_pred
This requires the backward propagation to be updated.
LinReg + Sigmoid = LogReg, so LinReg Layer + Sigmoid = LogReg Layer?
Throwing formulas around:
Updating our backward method:
def backward(self, inputs, outputs, alpha = 1e-5):
# Get the number of samples in dataset
m = inputs.shape[0]
# Forward propagate
Z1 = np.matmul(inputs, self.W1)
Z1_b = Z1 + self.b1
A1 = self.sigmoid(Z1_b)
Z2 = np.matmul(A1, self.W2)
Z2_b = Z2 + self.b2
A2 = self.sigmoid(Z2_b)
# Compute error term
dL_dA2 = -outputs/A2 + (1 - outputs)/(1 - A2)
dL_dZ2 = dL_dA2*A2*(1 - A2)
dL_dA1 = np.dot(dL_dZ2, self.W2.T)
dL_dZ1 = dL_dA1*A1*(1 - A1)
# Gradient descent update rules
self.W2 -= (1/m)*alpha*np.dot(A1.T, dL_dZ2)
self.W1 -= (1/m)*alpha*np.dot(inputs.T, dL_dZ1)
self.b2 -= (1/m)*alpha*np.sum(dL_dZ2, axis = 0, keepdims = True)
self.b1 -= (1/m)*alpha*np.sum(dL_dZ1, axis = 0, keepdims = True)
# Update Loss
self.CE_loss(inputs, outputs)
We use Activation functions to introduce non-linearity into our models. Sigmoid and tanh functions are often used, but both often lead to vanishing gradients.
Hence, we can use ReLU function or Leaky ReLU function due to their computational efficiencies and prevention of saturation
Improving gradient descent
We can improve gradient descent further (see Gradient Descent#Improving gradient descent) by introducing Momentum, Gradient-based learning rate control and Learning rate decay through different optimisation algorithms
Examples of optimisers
- AdaGrad
- RMSProp
- Adam optimiser
- AdaBound - Adam + AdaGrad
- AMSGrad - Improved Adam
- Lookahead - Adam + Momentum
Improving computation rate
We can perform parameter training updates with smaller batches of the dataset rather than the entire dataset per iteration by utilising Stochastic Gradient Descent, ideally mini-batch.
Good Practices
Additional performance metrics
Low loss does not mean successful training, because we have only minimised it. We should instead use more interpretable performance metrics
For instance, using accuracy:
def accuracy(self, inputs, outputs):
# Calculate accuracy for given inputs and ouputs
pred = [int(val >= 0.5) for val in self.forward(inputs)]
acc = sum([int(val1 == val2[0]) for val1, val2 in zip(pred, outputs)])/outputs.shape[0]
return acc
def train(self, inputs, outputs, N_max = 1000, alpha = 1e-5, beta1 = 0.9, beta2 = 0.999, \
delta = 1e-5, batch_size = 100, display = True):
# Get number of samples
M = inputs.shape[0]
# List of losses, starts with the current loss
self.losses_list = [self.CE_loss(inputs, outputs)]
self.accuracies_list = [self.accuracy(inputs, outputs)]
Train-test-validation split
We are not here to minimise the training loss function (or maximise accuracy) but to generalise well on unseen data. So expanding on Train and test split, we introduce a validation dataset.
This will help us access the performance of the model and how well it would generalise.
- Spotting Overfitting and Underfitting
- Choosing best Hyperparameters
Implementing early stop
In general,
- Model will start by underfitting the data, which is normal.
- After a few rounds of training models will (hopefully) achieve good generalization.
- Then, if training is pursued, the model will often attempt to minimize loss at all costs, often sacrificing generalisation in the process, and overfitting.
We should try and stop the training when the model starts losing it generalization capabilities and starts overfitting, when:
- Validation loss is minimal
- Validation and training curves start going in opposite ways
Early stopping can be defined by interrupting training when:
- A sufficiently high accuracy has been obtained (e.g. 98%)
- Accuracy is no longer increasing as the change between two consecutive iterations of accuracy falls below a threshold
Saver and Loader functions
Since we will often stop training after it is too late (after it loses generalisation capabilities), we should save model parameters every
def save(self, path_to_file, iter_num = "final"):
# Display
folder = path_to_file + "/" + iter_num + "/"
print("Saving model to", folder)
# Check if directory exists
if(not os.path.exists(folder)):
os.mkdir(folder)
# Dump
with open(folder + "W1.pkl", 'wb') as f:
pickle.dump(self.W1, f)
f.close()
with open(folder + "W2.pkl", 'wb') as f:
pickle.dump(self.W2, f)
f.close()
with open(folder + "b1.pkl", 'wb') as f:
pickle.dump(self.b1, f)
f.close()
with open(folder + "b2.pkl", 'wb') as f:
pickle.dump(self.b2, f)
f.close()
# Save model
self.save("./save", iter_num = str(iteration_number))
After training, load the model parameters (
Saving is also used to ensure reproducibility of results and preventing data loss.
Dense Neural Networks
A neural network with more than two hidden layers
Good practices
- Size of layers should progressively decrease by a factor of at least 2 e.g.
- Create building blocks for modularity
class DenseReLU(torch.nn.Module):
def __init__(self, n_x, n_y):
super().__init__()
# Define Linear layer using the nn.Linear()
self.fc = torch.nn.Linear(n_x, n_y)
def forward(self, x):
# Wx + b operation
# Using ReLU operation as activation after
return torch.relu(self.fc(x))
class DenseNoReLU(torch.nn.Module):
def __init__(self, n_x, n_y):
super().__init__()
# Define Linear layer using the nn.Linear()
self.fc = torch.nn.Linear(n_x, n_y)
def forward(self, x):
# Wx + b operation
# No activation function
return self.fc(x)
- Assemble the building blocks into a large Deep NN object
class DeepNeuralNet(torch.nn.Module):
def __init__(self, n_x, n_h, n_y):
super().__init__()
# Define three Dense + ReLU Layers,
# followed by one Dense + Softmax Layer
self.layer1 = DenseReLU(n_x, n_h[0])
self.layer2 = DenseReLU(n_h[0], n_h[1])
self.layer3 = DenseReLU(n_h[1], n_h[2])
self.layer4 = DenseNoReLU(n_h[2], n_y)
# Combine all four layers
self.combined_layers = torch.nn.Sequential(self.layer1,
self.layer2,
self.layer3,
self.layer4)
def forward(self, x):
# Flatten images (transform them from 28x28
# 2D matrices to 784 1D vectors)
x = x.view(x.size(0), -1)
# Pass through all four layers
out = self.combined_layers(x)
return out
# Initialize the model and optimizer
model = DeepNeuralNet(n_x=784, n_h=[80, 40, 20], n_y=10).to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3)
Deeper networks may be prone to overfitting if the network is too deep.
Conversely, shallower networks may be easier to train and require less data but may not be able to learn as complex patterns.